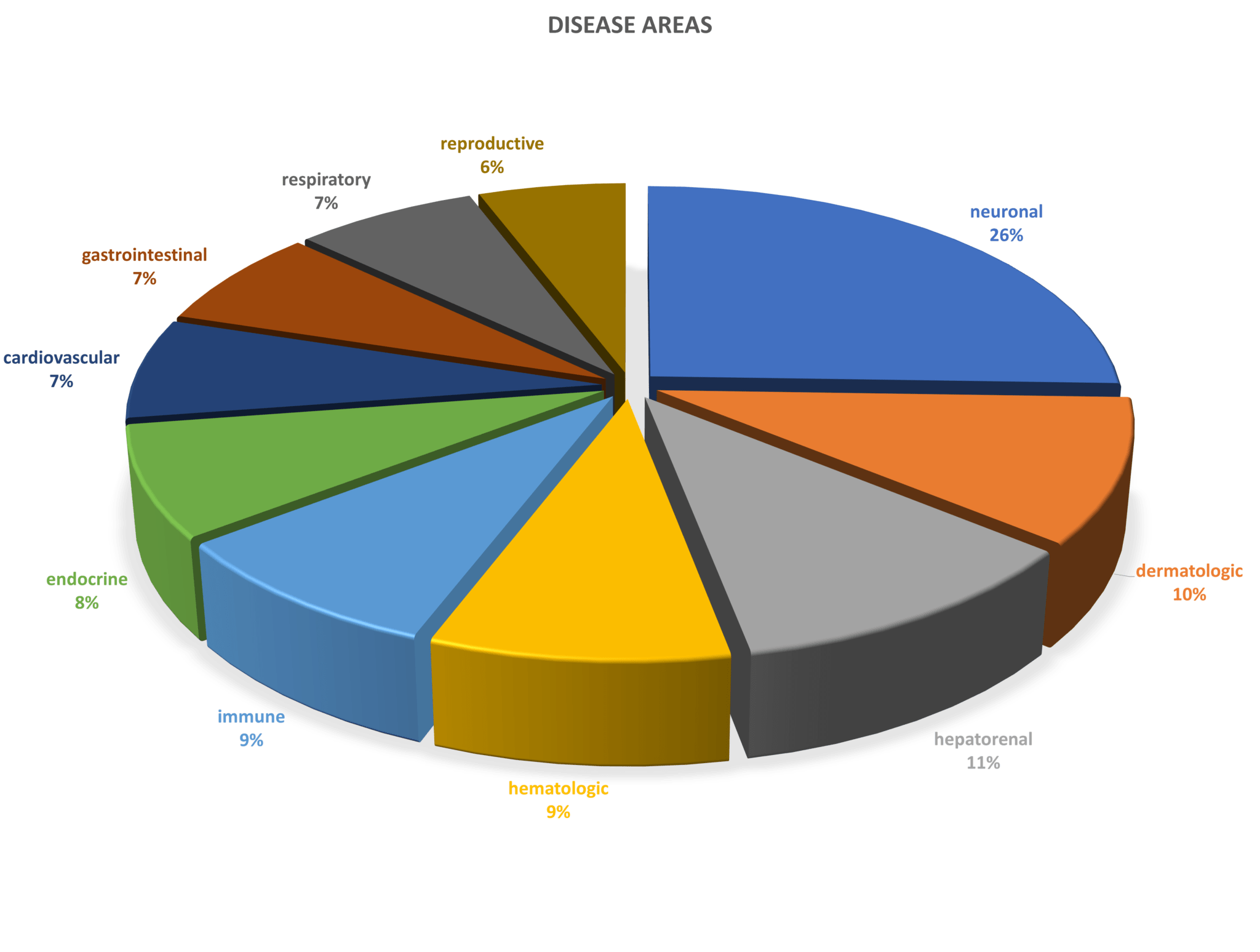
SML Meditree is a leading MFDS-certified GCLP institution in Seoul, South Korea that provides a variety of clinical sample analyses services to pharmaceutical companies, bio-ventures, food industries, and researchers to help advance their clinical trials and research projects. They provide high-quality analyses and rapid services to accelerate new drug developments worldwide. With Sengenics new proteomics technology, they will be able to provide additional proteomics services to assist with biomarker discovery, patient stratification strategies for clinical research and therapeutics, and outcome predictions.
Learn more about SML Meditree
SML메디트리 (medi-tree.com)